Most basic
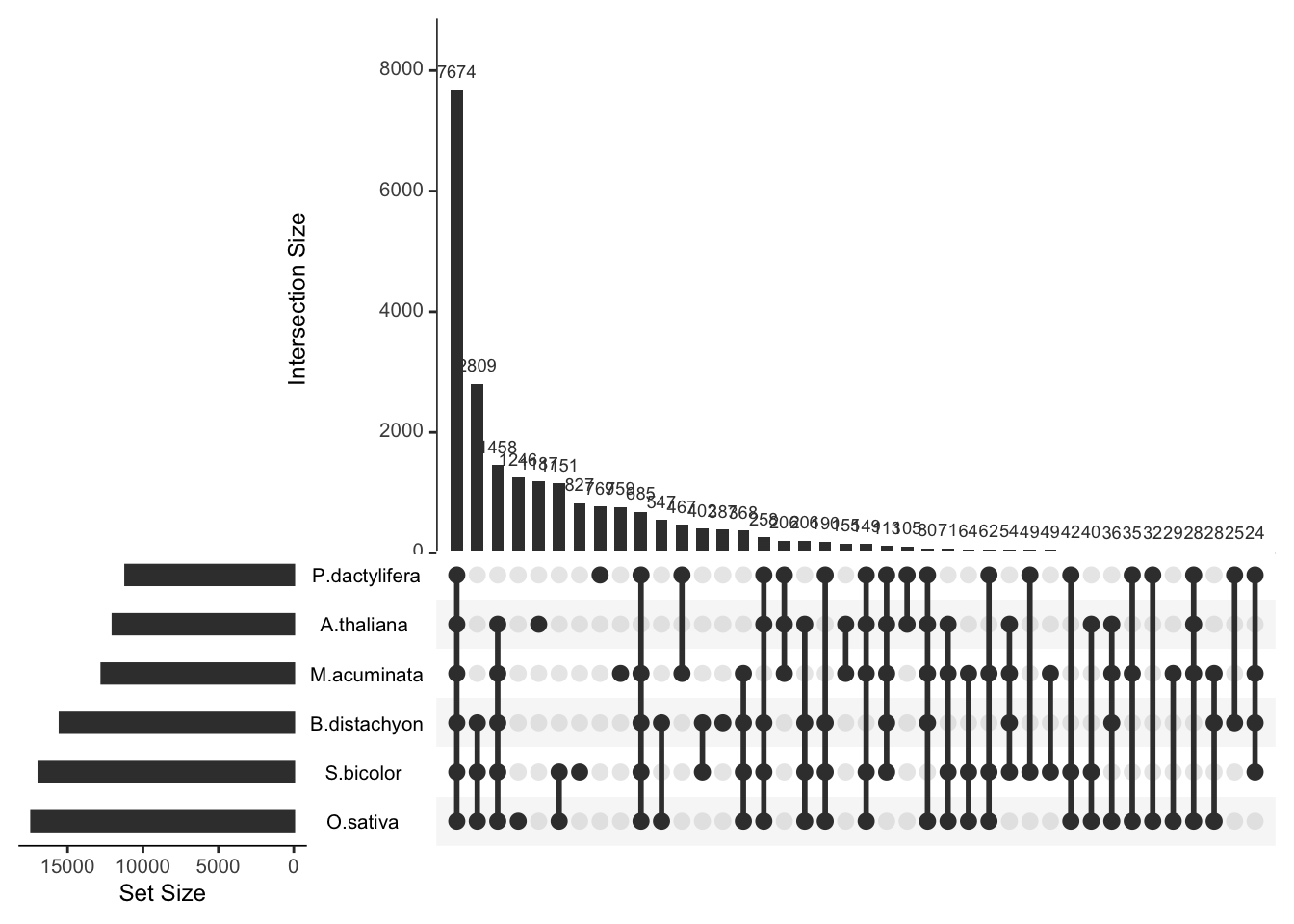
To visualize the intersection between more than 3 sets, the best option is to use an UpSet plot.
Here is an example provided by the UpsetR package that
displays information concerning the Banana genome.
The total size of each set is represented on the left barplot. Every possible intersection is represented by the bottom plot, and their occurence is shown on the top barplot.
Source: the upSetR documentation
# Specific library
library(UpSetR)
# Dataset
input <- c(
M.acuminata = 759,
P.dactylifera = 769,
A.thaliana = 1187,
O.sativa = 1246,
S.bicolor = 827,
B.distachyon = 387,
"P.dactylifera&M.acuminata" = 467,
"O.sativa&M.acuminata" = 29,
"A.thaliana&O.sativa" = 6,
"S.bicolor&A.thaliana" = 9,
"O.sativa&P.dactylifera" = 32,
"S.bicolor&P.dactylifera" = 49,
"S.bicolor&M.acuminata" = 49,
"B.distachyon&O.sativa" = 547,
"S.bicolor&O.sativa" = 1151,
"B.distachyon&A.thaliana" = 10,
"B.distachyon&M.acuminata" = 9,
"B.distachyon&S.bicolor" = 402,
"M.acuminata&A.thaliana" = 155,
"A.thaliana&P.dactylifera" = 105,
"B.distachyon&P.dactylifera" = 25,
"S.bicolor&O.sativa&P.dactylifera" = 42,
"B.distachyon&O.sativa&P.dactylifera" = 12,
"S.bicolor&O.sativa&B.distachyon" = 2809,
"B.distachyon&O.sativa&A.thaliana" = 18,
"S.bicolor&O.sativa&A.thaliana" = 40,
"S.bicolor&B.distachyon&A.thaliana" = 14,
"O.sativa&B.distachyon&M.acuminata" = 28,
"S.bicolor&B.distachyon&M.acuminata" = 13,
"O.sativa&M.acuminata&P.dactylifera" = 35,
"M.acuminata&S.bicolor&A.thaliana" = 21,
"B.distachyon&M.acuminata&A.thaliana" = 7,
"O.sativa&M.acuminata&A.thaliana" = 13,
"M.acuminata&P.dactylifera&A.thaliana" = 206,
"P.dactylifera&A.thaliana&S.bicolor" = 4,
"O.sativa&A.thaliana&P.dactylifera" = 6,
"S.bicolor&O.sativa&M.acuminata" = 64,
"S.bicolor&M.acuminata&P.dactylifera" = 19,
"B.distachyon&A.thaliana&P.dactylifera" = 3,
"B.distachyon&M.acuminata&P.dactylifera" = 12,
"B.distachyon&S.bicolor&P.dactylifera" = 23,
"M.acuminata&B.distachyon&S.bicolor&A.thaliana" = 54,
"P.dactylifera&S.bicolor&O.sativa&M.acuminata" = 62,
"B.distachyon&O.sativa&M.acuminata&P.dactylifera" = 18,
"S.bicolor&B.distachyon&O.sativa&A.thaliana" = 206,
"B.distachyon&M.acuminata&O.sativa&A.thaliana" = 29,
"O.sativa&M.acuminata&A.thaliana&S.bicolor" = 71,
"M.acuminata&O.sativa&P.dactylifera&A.thaliana" = 28,
"B.distachyon&M.acuminata&O.sativa&A.thaliana" = 7,
"B.distachyon&S.bicolor&P.dactylifera&A.thaliana" = 11,
"B.distachyon&O.sativa&P.dactylifera&A.thaliana" = 5,
"A.thaliana&P.dactylifera&S.bicolor&O.sativa" = 21,
"M.acuminata&S.bicolor&P.dactylifera&A.thaliana" = 23,
"M.acuminata&B.distachyon&S.bicolor&P.dactylifera" = 24,
"M.acuminata&O.sativa&S.bicolor&B.distachyon" = 368,
"P.dactylifera&B.distachyon&S.bicolor&O.sativa" = 190,
"P.dactylifera&B.distachyon&S.bicolor&O.sativa&A.thaliana" = 258,
"P.dactylifera&M.acuminata&S.bicolor&B.distachyon&O.sativa" = 685,
"M.acuminata&S.bicolor&B.distachyon&O.sativa&A.thaliana" = 1458,
"S.bicolor&M.acuminata&P.dactylifera&O.sativa&A.thaliana" = 149,
"B.distachyon&M.acuminata&P.dactylifera&O.sativa&A.thaliana" = 80,
"M.acuminata&S.bicolor&B.distachyon&P.dactylifera&A.thaliana" = 113,
"M.acuminata&S.bicolor&B.distachyon&P.dactylifera&O.sativa&A.thaliana" = 7674
)
# Plot
upset(fromExpression(input),
nintersects = 40,
nsets = 6,
order.by = "freq",
decreasing = T,
mb.ratio = c(0.6, 0.4),
number.angles = 0,
text.scale = 1.1,
point.size = 2.8,
line.size = 1
)